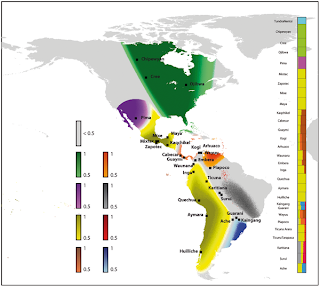
An open access PLOS article directing a computer to cluster Native Americans into seven clusters, further informed by language and geographical information produced the map above.
Previous analyses comparing genetic to linguistic differentiation in the Americas yielded equivocal results. Cavalli-Sforza et al. reported that, prior to the publication of their book, three of seven studies supported congruence between genes and languages. At that time, Ward et al. found that rates of linguistic diversification are faster that rates of genetic differentiation in mtDNA, and concluded that there is little congruence between linguistic and genetic relationships in the Americas. In more recent studies also using mtDNA, the hypothesis that language classifications reflect the genetic structure of Native American populations was also rejected. Lastly, an analysis of autosomal microsatellite markers in 28 Native American populations from the Human Genome Diversity Panel (HGDP) provided a qualitative correspondence between linguistic and genetic groupings. However, tests of correlation were not significant for these data.
In this study, linguistic data did corollate with autosomal genetics, something that the authors attribute to the fact that the latent cluster methodology that they used does not require any particular tree-like model to be generated. The linguistic data input in variants of the models used were as follows: "Model B used Greenberg's classification at the stock level (8 levels), Model C used Greenberg's classification at the group level (14 levels), and Model D used The Ethnologue classification at the family level (16 levels)."
Unsurprisingly, Ethnologue's more details account provided a better match to the genetic clusters. "Among the 16 families of The Ethnologue classification, only the Tupi, Choco and Chibchan families were not associated to a unique genetic cluster." The Choco (Waunana and Embera) and Chibchan (Cabacar, Guayme, Kogi and Arhuaco)correspond to the populations in the brownish-orange colored area that includes most of Central America and the northern coast of South America. The Tupi languages (Guarani, Karbana, Ache and Surui) include the three groups on the map in the general vicinity of Uruguay, and Surui in the vicinity of Bolivia.
Put another way, the yellow (Oto-Manguean Mixtec and Zapotec, Mixe-Zoque Mixe, Mayan Maya and Kaqchikel, Quechuan Inga and Quechua, Amaryan Amarya, Auraucanian Hulliche, Marco-Ge Kaingang, Arawakan Wayuu and Piapoco), green (Na-Dene Chipawyan, and Algic Cree and Ojibwa) and purple (Uto-Aztecan Pima) clusters were cases where every language cluster in the group fit entirely into a single genetic cluster, while the language groups outside those areas did not fit a single genetic cluster. The number of clusters chosen was arbitrary.
Analysis
While the authors don't articulate their conclusions in these terms, the implication of a situation where there are overlapping genetic and linguistic clusters that lack a tree-like structure seems to be that Native American languages may have been part of various Sprachbunds, rather than having a tree-like relationship to each other. This would be consistent with a scenario in which Native Americans speaking a very small number of common languages (perhaps just one or two) disperse rapidly across the Americas, and then remain more or less geographically fixed in place as their languages begin to random drift apart from each other influenced by neighbors.
An alternate interpretation, for the yellow regions at least, each of which were parts of Pre-Columbian Neolithic civilizations (the Incas on the Pacific Coast of South America, and successions of Olmecs, Mayans and Atzecs in the general vicinity of Mexico), is that the two Neolithic populations were fairly close genetically and that their Neolithic expansions created the genetic cluster, but did not extend to the rest of Central America and South America, which was unsuitable for their crops, and hence allowed the greater pre-Neolithic genetic diversity of Latin America to persist. The purple Uto-Aztecan cluster could have a similar source.
Perhaps also, the Central American and South American regions that do not cluster so neatly have a mix of settlers who came from the Andes and went East, and settlers who made their way along the Atlantic coast and forged inland.
In contrast, the green areas in North America may have had only a single wave of initial settlement that was relatively distinct from the Latin American regions; other evidence puts the Eastern and Western branches of Native Americans as the deepest genetic divide in a generally genetically unified population derived from a small founding population.
An earlier post at this blog on the same topic is found here.
2 comments:
I could only find this older paper which essentially produces the same results (with less pretense probably). However I recall a paper that reached down to K=16 or something like that and certainly that did split the Mesoamerican-Andean cluster, which may be an illusion to some extent.
Thanks for blogging this. 2 thoughts: 1) a well-studied sprachbund is the Northern California/Southern Oregon sprachbund at the boundary between some Penutian and Hokan languages (Scott Delancey at U of Oregon sutides this) which lands in one of the non-matching areas - no surprise that the major "port of entry" into the Americas, i.e. the northwestern quarter of North America, should be messier than the rest. Surprising to me is that there are any Na-Dene languages matching Algic, although in that part of Canada (central/western boreal forest) there have probably been several millennia of contact between Algic and Na-Dene speakers, and plenty of opportunity for gene flow.
Post a Comment